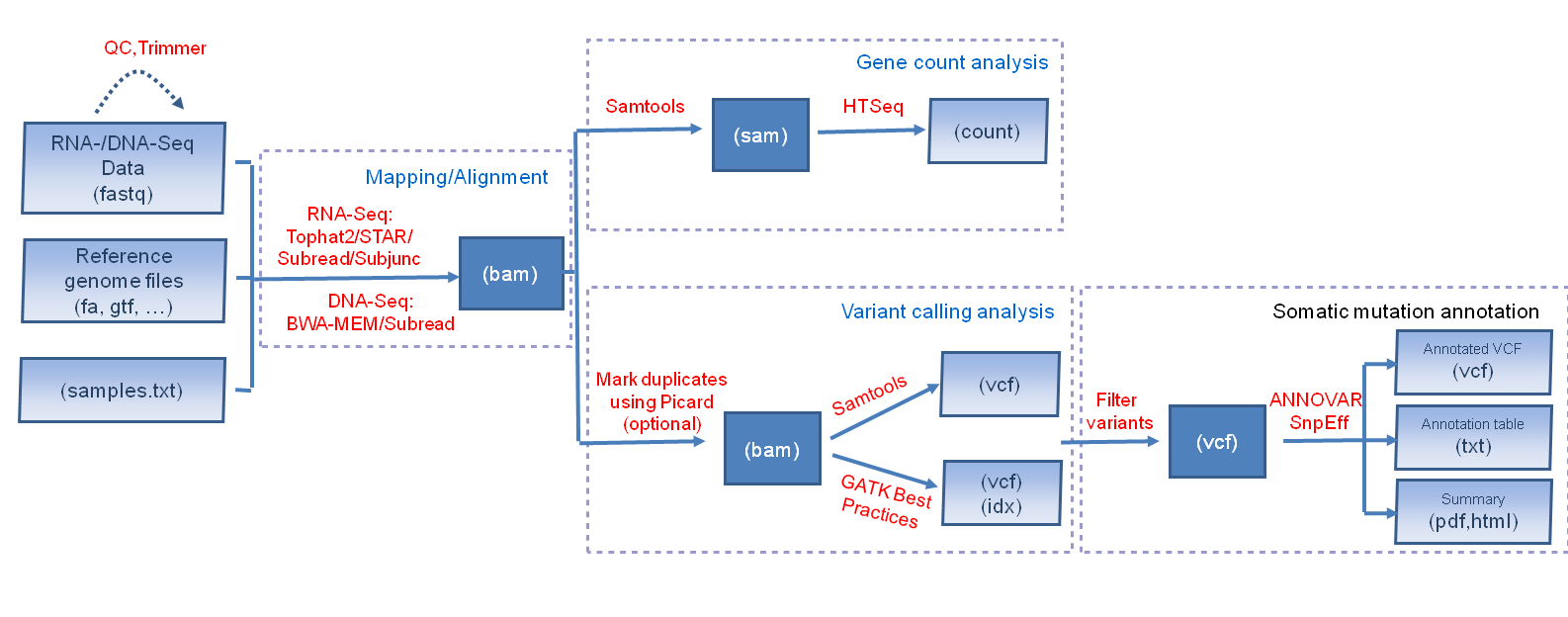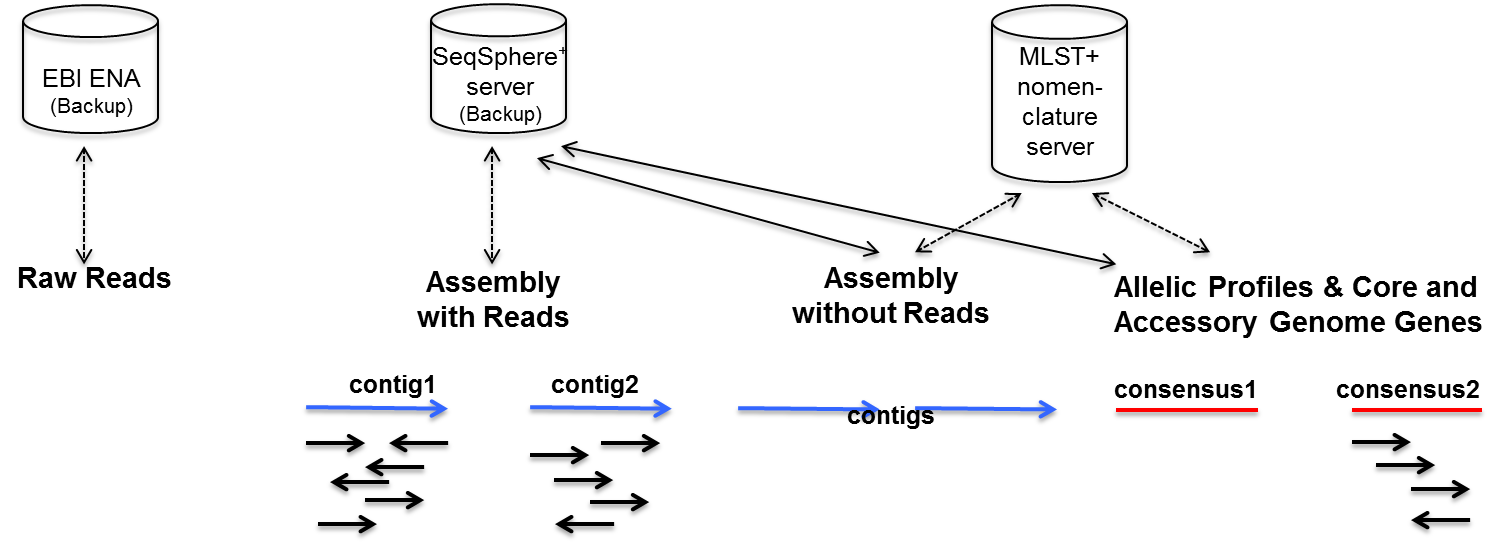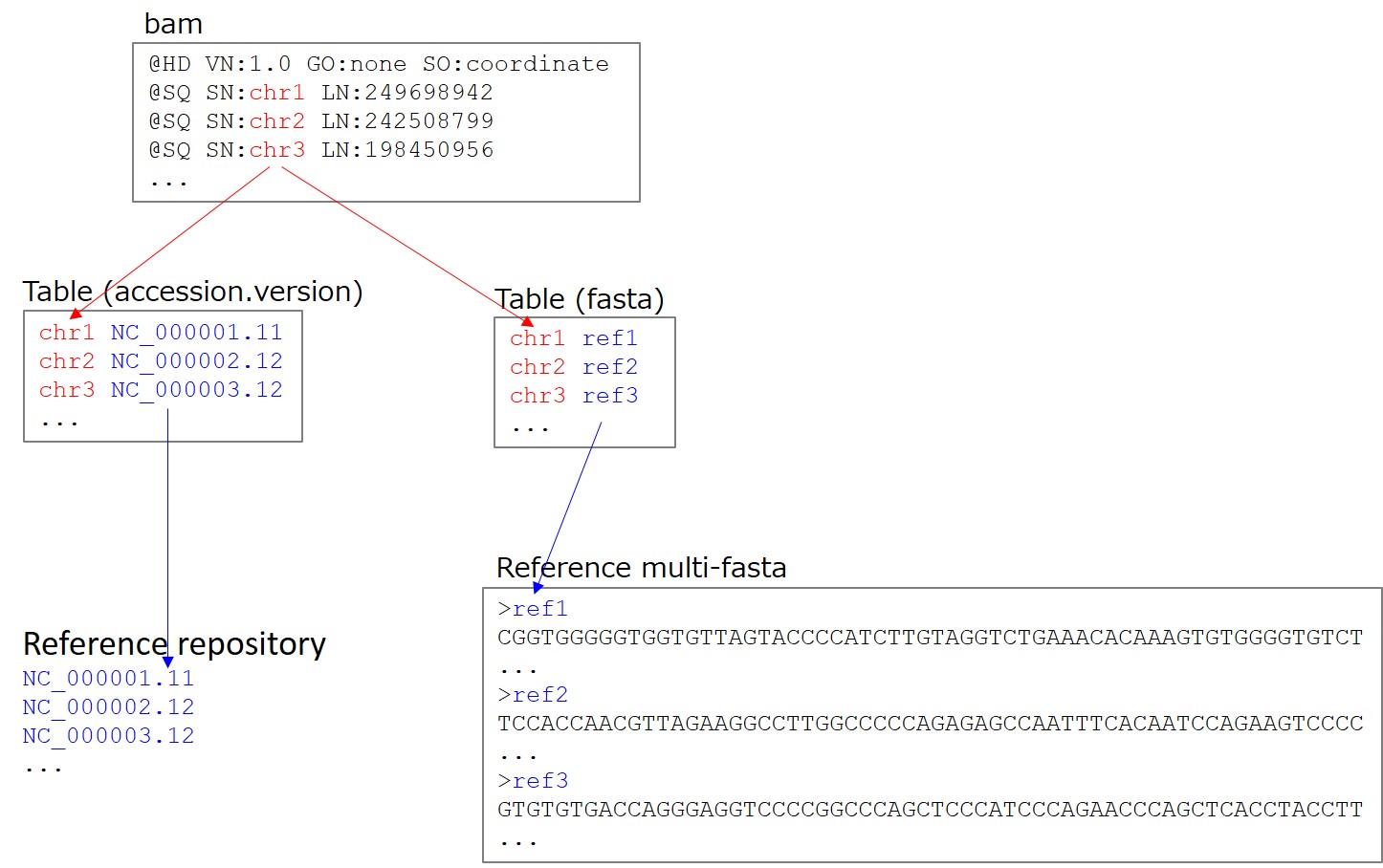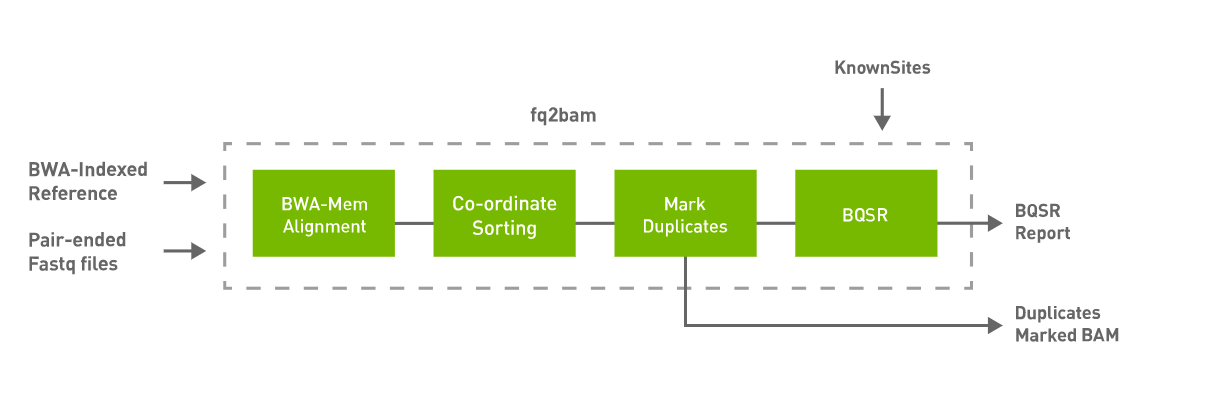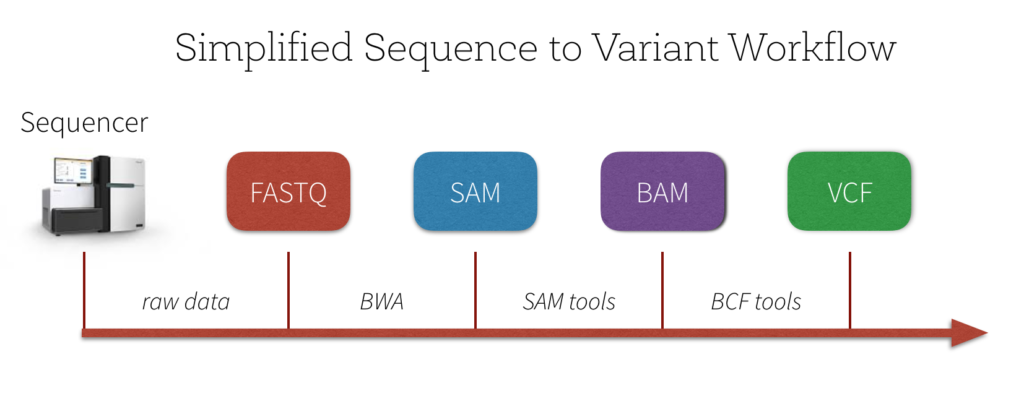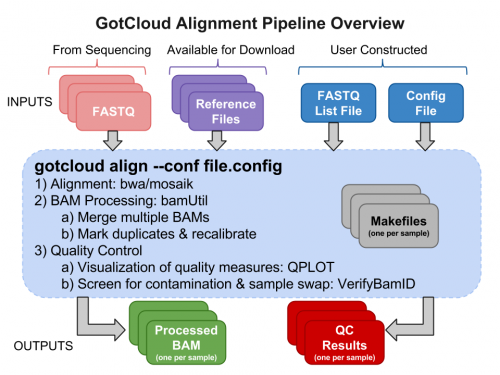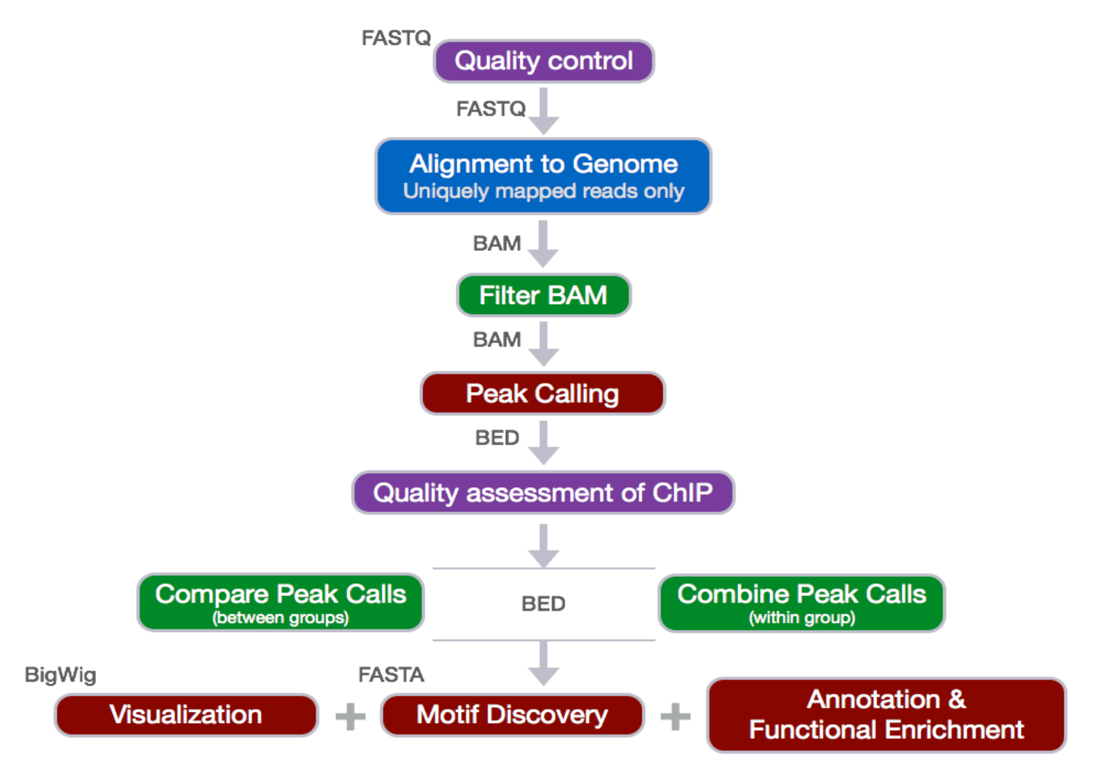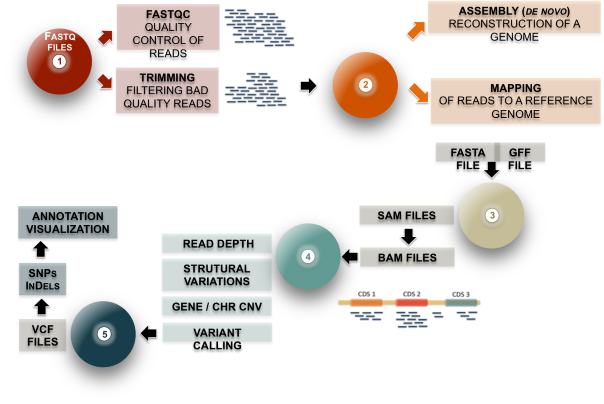
3. Day 2: Introduction to basic NGS pipelines — CODATA-RDA Research Data Science Advanced Workshop on Bioinformatics 1.0 documentation

FastGT: an alignment-free method for calling common SNVs directly from raw sequencing reads | Scientific Reports
GitHub - PacificBiosciences/bam2fastx: Converting and demultiplexing of PacBio BAM files into gzipped fasta and fastq files.